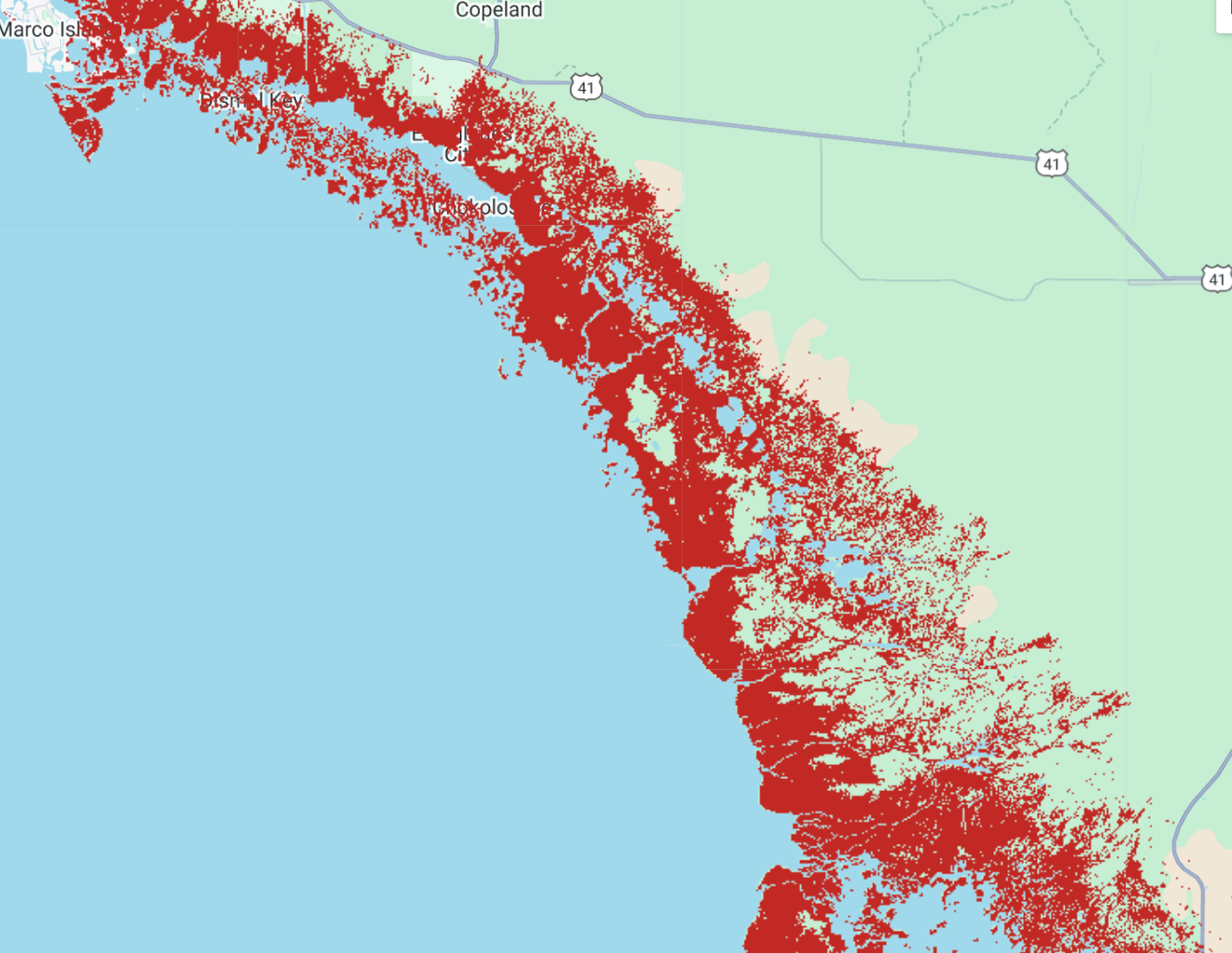
An attempt to fine-tune Prithvi-EO to produce mangrove classification maps.
This work is based on previous post Seagrass Mapping with NASA Prithvi-EO.
Code related to this post is stored in 7yl4r/nasa-prithvi-wetlands.
Region of Interest (RoI)
The Ten Thousand Islands (TTI) area is chosen for assessment of mangrove classification because of * overlapping data from the RBNERR Mangrove Coast Collective project * RBNERR interest in mangrove assessment * major mangrove impacts from 2022 hurricane Ian
Input Files
Spectral Training Input
A sentinel-2 spectral median was prepared for the RoI. This GEE script was used to generate the file.
The CloudScorePlus product was used with a threshold of 0.80 to filter clouds. The recommended value for the threshold is 0.65, but for this region the 0.8 was required to remove visable hazes in the median product.
Images from years 2023-2025 were used in three distinct seasons.
High Attenuation (Jan-Apr) - Image count:
663
Transitional (May-early Jul) - Image count:
439
Peak Visibility (Late Jul-Oct) - Image count:
642The file is placed in data/inputs/spectral/TTI_seasonal_s2_stack.tif.
Mangrove Classification Map Training Input
The LANDSAT/MANGROVE_FORESTS layer produced by Giri et alii from GEE was used. This GEE script was used to export the file data/inputs/classmaps/TTI_mangrove_landsat_Giri.tif.
Evaluation Spectral Image
An image was prepared for the tuned classifier to be used upon, TTI_seasonal_s2_stack_2019_to_2021.tif.
Methods matching the Spectral Training Input file above were used for the years 2019-2021.
High Attenuation (Jan-Apr) - Image count:
673
Transitional (May-early Jul) - Image count:
408
Peak Visibility (Late Jul-Oct) - Image count:
581The file was uploaded to GDrive for access from colab notebooks.
Methods
Preparation of Fine-Tuning Patch Chips
The input files were downloaded and a script was used to cut these images into into sets of chips:
(base) tylar@tylar-laptop:~/repos/nasa-prithvi-wetlands$ python scripts/extract_tuning_patches_mangrove_sentinel2.py
Clearing existing output directory: data/output/tuning_patches
Detected single TIF file: data/input/spectral/TTI_seasonal_s2_stack.tif
============================================================
Processing spectral file 1/1: TTI_seasonal_s2_stack.tif
============================================================
Spectral CRS: EPSG:4326
Mask CRS: EPSG:4326
Spectral bounds (native): BoundingBox(left=-81.90290788278485, bottom=25.280748051805226, right=-80.7931291807836, top=26.05823993021067)
Mask bounds (native): BoundingBox(left=-81.90290788278485, bottom=25.280748051805226, right=-80.7931291807836, top=26.05823993021067)
Overlap bounds (in spectral CRS): left=-81.90, bottom=25.28, right=-80.79, top=26.06
Spectral file shape: (2885, 4118)
Mask file shape: (2885, 4118)
Overlap region: 2885x4118 pixels
Spectral window: Window(col_off=0, row_off=0, width=4118, height=2885)
Mask window: Window(col_off=0, row_off=0, width=4118, height=2885)
Extracting 224x224 patches with stride 224
Number of spectral bands: 24
Will extract up to 216 patches from this shard
Extracted 100 total valid patches...
Extracted 200 total valid patches...
Shard complete: 216 valid patches extracted
============================================================
Extraction complete!
============================================================
Spectral files processed: 1
Valid patches saved: 216
Patches skipped: 0
Output directory: data/output/tuning_patches
Organizing patches into training and validation sets...
Dataset split created:
Training samples: 173
- Files in: data/output/tuning_patches/training_chips
- List file: training_data.txt
Validation samples: 43
- Files in: data/output/tuning_patches/validation_chips
- List file: validation_data.txt
Chip naming format:
- Spectral: chip_XXXXX_merged.tif
- Mask: chip_XXXXX.mask.tif
Cleaning up temporary directories...
- Removed: data/output/tuning_patches/spectral
- Removed: data/output/tuning_patches/masks
✓ Cleanup complete!
✓ Patch extraction complete! Ready for Prithvi fine-tuning.
Compressing tuning patches...
✓ Compressed patches saved to: data/output/mangrove_tuning_patches.tar.bz2The file was uploaded to gdrive for the ipnb to use.
Fine Tuning Research Notebook
The research notebook was developed based on the previous seagrass notebook was created.
Among the adjustments needed were: * adjusted filenames * adjusted band names, numbers, etc * adjusted class names, labels, etc
The final notebook was lost due to a computer crash, but an early iteration is here.
Results
The classification is reasonable but lowered resolution when compared to the input product.